An Exercise in Cleaning and Exploring NPDES Data for Permit Development
Introduction
This is an example exercise in cleaning and preparing NPDES data for use in a reasonable potential analysis using R. When cleaning data for an RPA, the goal is to pare away data which is not needed and to identify & correct irregularities (missing data, inconsistent labeling, etc) within what remains.
The point of this exercise is two-fold:
Providing a general NPDES data cleaning walkthrough for new permit writers.
Give some detail on how to tackle specific tasks using R. In my experience, most permit writers use spreadsheet tools for data cleaning so there are fewer resources to draw on for other tools.
The particular steps taken in any given data cleaning job will change depending on the content and format of the NPDES data in question. As a consequence, I find defining a set of hard and fast steps or rules for data cleaning is not helpful. Instead, a permit writer needs to get a sense for what a clean dataset ready for input to an RPA looks like, then map a path from that messy data to the desired endpoint.
Walkthrough
As a new permit writer, figuring out how to tackle data cleaning can be challenging–especially when one’s understanding of the RPA process itself is fuzzy. This document can serve as a starting place for planning and executing a data cleaning task. While the particulars may change from permit to permit, the general flow of this walkthrough (if not the exact steps) will be widely applicable.
Obtain NPDES Data
First, let’s use the new package ‘echor’ to download discharge monitoring data from the Hangtown Creek WRF, owned and operated by the City of Placerville. The package ‘echor’ provides an interface for connecting to EPA’s ECHO database and downloading data. We will use ‘dplyr’ for manipulating the dataset and ggplot2 for making graphs.
library(echor)
library(dplyr, warn.conflicts = FALSE)
library(ggplot2, warn.conflicts = FALSE)We will use the ‘echoGetEffluent’ function and the NPDES ID for the facility (CA0078956) to download the effluent data available in ECHO. We’ll specify that we want all data collected between January 2015 - December 2019. For those who want to follow along at home, you can use the code immediately below to download the data I’ll be using. I’ve included paginated copies of the dataset at key places below, but its inherently awkward to flip through the data on a webpage so it may be easier explore the data in your own R session.
htc <- echor::echoGetEffluent("CA0078956",
start_date = "01/01/2015",
end_date = "12/31/2019")
saved <- htc
write.csv(saved, file = "CA0078956_raw_data.csv")
htcNote that after the data was downloaded we saved a copy of the raw data to the computer in a .csv file named “CA0078956_raw_data.csv”. It is good practice to save a copy of your raw, unedited data before cleaning. This is true whether you are using R, Python, or Excel. Having a copy of the raw data gives analysts and quality control auditors something to refer back to when checking the steps of the RPA and limit calculations.
Cleaning the Dataset
Now that we have the data, let’s look at what’s inside. Let’s list the column headers, the pollutant parameters included in the dataset, and the monitoring locations where the data was collected.
headers <- names(htc)
headers## [1] "activity_id" "npdes_id"
## [3] "version_nmbr" "perm_feature_id"
## [5] "perm_feature_nmbr" "perm_feature_type_code"
## [7] "perm_feature_type_desc" "limit_set_id"
## [9] "limit_set_schedule_id" "limit_id"
## [11] "limit_type_code" "limit_begin_date"
## [13] "limit_end_date" "nmbr_of_submission"
## [15] "parameter_code" "parameter_desc"
## [17] "monitoring_location_code" "monitoring_location_desc"
## [19] "stay_type_code" "stay_type_desc"
## [21] "limit_value_id" "limit_value_type_code"
## [23] "limit_value_type_desc" "limit_value_nmbr"
## [25] "limit_unit_code" "limit_unit_desc"
## [27] "standard_unit_code" "standard_unit_desc"
## [29] "limit_value_standard_units" "statistical_base_code"
## [31] "statistical_base_short_desc" "statistical_base_type_code"
## [33] "statistical_base_type_desc" "limit_value_qualifier_code"
## [35] "stay_value_nmbr" "dmr_event_id"
## [37] "monitoring_period_end_date" "dmr_form_value_id"
## [39] "value_type_code" "value_type_desc"
## [41] "dmr_value_id" "dmr_value_nmbr"
## [43] "dmr_unit_code" "dmr_unit_desc"
## [45] "dmr_value_standard_units" "dmr_value_qualifier_code"
## [47] "value_received_date" "days_late"
## [49] "nodi_code" "nodi_desc"
## [51] "exceedence_pct" "npdes_violation_id"
## [53] "violation_code" "violation_desc"
## [55] "rnc_detection_code" "rnc_detection_desc"
## [57] "rnc_detection_date" "rnc_resolution_code"
## [59] "rnc_resolution_desc" "rnc_resolution_date"
## [61] "violation_severity"That’s a lot of columns! Let’s see if we can narrow the list down. We’ll keep information pertaining to the monitoring location, the parameter identity, when the discharge occurred, and the parameter measurement information.
columns_keep <- c("perm_feature_nmbr", "perm_feature_type_desc",
"parameter_code", "parameter_desc",
"monitoring_period_end_date", "dmr_value_nmbr",
"dmr_unit_desc", "dmr_value_qualifier_code",
"statistical_base_short_desc",
"nodi_code", "nodi_desc")
htc <- htc[ , columns_keep]htc %>%
group_by(perm_feature_nmbr, perm_feature_type_desc) %>%
summarise(n = n())Based on the table above, it appears that the ECHO database includes influent and effluent data but no receiving water data for this plant. In this type of circumstance, I’d examine the NDPES permit–was the discharger supposed to collect receiving water data? If they were, then I would double-check my database query to make sure I hadn’t somehow missed it (maybe widen the date range I searched under). If the query looked okay, then I would ask the discharger about it.
Let’s filter out the influent data since its not used in the RPA and limit our exercise to solely effluent data. Then we will examine the list of parameters in the dataset.
htc <- filter(htc, perm_feature_nmbr == "001")htc$parameter_desc %>%
unique() %>%
sort() %>%
data.frame(parameter_name = .)We can use the table above to flip through all the parameters included in the dataset. In a real data cleaning task, we would start pruning this list down to the parameters we wish to include in the RPA. To figure out what those are, we would make reference to the water quality objectives and water quality standard implementing procedures effective in the permittee’s jurisdiction. To make this task quicker, I typically maintain a table of pollutants of concern which are applicable to a jurisdiction or to a class of dischargers (e.g., POTWs). I will do a join of the dataset with the pollutant-of-concern table, and discard parameters which are not pollutants of concern.
Before pruning the list down to formal pollutants of concern for the RPA, it may be advisable to edit the parameter names to match some standardized format. This will make joining/matching the tables more accurate. In addition, the parameters may not be named in an internally consisitent way–e.g., the zinc entries may be labeled “Zinc, total recoverable” in some cases, and “Total Zinc” in others. Finally, there may be other reasons for standardizing pollutant names. For example, it may be important that zinc be labeled “Zinc” instead of “Zinc, total recoverable” or “Total Zinc” in whatever RPA calculation tool which will be used to process the clean data.
For this exercise, we will delay pruning the pollutant list so as to better illustrate subsequent cleaning steps. We will select the list of pollutants of concern at the end of the exercise prior to summarizing the data.
Next, let’s examine the structure of the data table.
str(htc)## tibble [2,536 x 11] (S3: tbl_df/tbl/data.frame)
## $ perm_feature_nmbr : chr [1:2536] "001" "001" "001" "001" ...
## $ perm_feature_type_desc : chr [1:2536] "External Outfall" "External Outfall" "External Outfall" "External Outfall" ...
## $ parameter_code : chr [1:2536] "00010" "00010" "00010" "00010" ...
## $ parameter_desc : chr [1:2536] "Temperature, water deg. centigrade" "Temperature, water deg. centigrade" "Temperature, water deg. centigrade" "Temperature, water deg. centigrade" ...
## $ monitoring_period_end_date : chr [1:2536] "01/31/2015" "02/28/2015" "03/31/2015" "04/30/2015" ...
## $ dmr_value_nmbr : chr [1:2536] "15.8" "17.2" "18.6" "17" ...
## $ dmr_unit_desc : chr [1:2536] "deg C" "deg C" "deg C" "deg C" ...
## $ dmr_value_qualifier_code : chr [1:2536] "=" "=" "=" "=" ...
## $ statistical_base_short_desc: chr [1:2536] "INST MAX" "INST MAX" "INST MAX" "INST MAX" ...
## $ nodi_code : chr [1:2536] "" "" "" "" ...
## $ nodi_desc : chr [1:2536] "" "" "" "" ...It appears that all columns were read in as character vectors so let’s convert the date and effluent measurement columns into date and numeric types, respectively. We will definitely need to be able to sort numbers later on, and being able to sort dates may come in handy when doing QC on RPA results.
htc$monitoring_period_end_date <- lubridate::mdy(htc$monitoring_period_end_date)
htc$dmr_value_nmbr <- as.numeric(htc$dmr_value_nmbr)Next, let’s look at the statistical base codes (i.e., the column ‘statistical_base_short_desc’). This column tells us if the reported value is a single-day measurement which will be listed as a daily max if the monitoring frequency is daily or less frequent. We are interested in the individual measurements and not statistical aggregations, like monthly or weekly averages. We’ll also look at units so we can distinguish between concentration data (e.g., mg/L) and mass flux data (lbs/day). RPAs are typically based on concentration values rather than mass fluxes.
htc %>%
group_by(parameter_desc) %>%
summarise(units = paste(dmr_unit_desc, collapse = ", "),
stat_desc = paste(statistical_base_short_desc, collapse = ", "))
htc <- htc %>%
filter(statistical_base_short_desc == "DAILY MX") htc %>%
select(parameter_desc, dmr_unit_desc, statistical_base_short_desc) %>%
group_by(parameter_desc) %>%
unique() %>%
summarise(units = paste(dmr_unit_desc, collapse = ", "),
stat_base = paste(statistical_base_short_desc, collapse = ", ")) %>%
rmarkdown::paged_table()htc <- htc %>%
filter(statistical_base_short_desc == "DAILY MX")Based on flipping through the data in the table above, it appears we want to keep the “DAILY MX” observations and drop the rest, which appear to correspond to non-toxics or to weekly/monthly aggregations.
The only toxics with mixed units (i.e., sometimes ug/L is used, other times ng/L) are mercury and methylmercury, so we will convert units such that everything is in ug/L. Normally, we would also compare the units used here to the units in the water quality objectives to ensure they are consistent, but we will skip that step for purposes of this exercise.
htc <- htc %>%
mutate(dmr_value_nmbr = case_when(dmr_unit_desc == "ng/L" ~ dmr_value_nmbr / 1000,
TRUE ~ dmr_value_nmbr),
dmr_unit_desc = case_when(dmr_unit_desc == "ng/L" ~ "ug/L",
TRUE ~ dmr_unit_desc))Let’s also remove the mass flux data (indicated by “lb/d” units), the fiber data (“Fib/L”), flow (“MGD”), conductance (“umho/cm”), and chronic toxicity (“tox chronic”). We’ll keep the mass concentration data and observations where they forgot to include a unit indicator.
htc <- htc %>%
filter(dmr_unit_desc %in% c("mg/L", "", "ug/L", "ng/L"))Next, let’s look at the data with missing units.
htc %>%
filter(dmr_unit_desc == "") It appears all of the observations with missing units belong to observations that were reported as unquantifiable. In a real job, we would reconcile this missing information with the permittee (i.e., obtain method detection limits, and reporting levels) if it appeared the missing information could influence reasonable potential findings or effluent limitation calculations.
So we started with 2896 observation rows and we’re down to 722. Next let’s see what date ranges the data covers.
htc %>%
group_by(parameter_desc) %>%
summarize(min = min(monitoring_period_end_date),
max = max(monitoring_period_end_date),
n_samples = n()) %>%
arrange(desc(n_samples))Based on this table, it looks like most of the toxics one would use in an RPA were monitored once (April 2017) during the period of interest. The exceptions are lead, mercury, and zinc. Perhaps ammonia, too, depending on the water quality standards.
At this point, we would typically compare our dataset to the permit monitoring requirements to ascertain whether we had all the data we were supposed to have. Are there pollutants the discharger should have monitored for which were not included in this dataset? And were all pollutants monitored the correct number of times–e.g., if the permit requires monthly monitoring for zinc then we should have 60 samples within the five years of data. For purposes of this example exercise we will skip this step, but its very important to do when preparing data for an RPA in the real world.
Next, let’s make sure that we have valid numbers for all of our toxics data. Were any observations left blank or marked NA? Were non-detect and non-quantifiable values listed with method detection limits and method reporting levels. If any missing information is identified, then we would need to contact the discharger to obtain any needed missing information.
First, let’s search for any result data listed as blank (e.g., "") or NA.
htc %>%
filter(dmr_value_nmbr == "" | is.na(dmr_value_nmbr)) As noted above with the missing units, It looks like we have a number of items marked as unquantifiable. If knowing the method detection limits/reporting levels for these items could conceivably change our RPA findings, or if it might change the effluent limit imposed, we would want to contact the discharger and collect the missing information.
Next, we will look at the qualified data and see if we have detection limit information. While we are at it, we’ll also make it so qualifiers are consistently applied (i.e., all detect values will be marked " “, instead of a mix of”=" signs and blank " " values).
htc[which(htc$dmr_value_qualifier_code == "="), "dmr_value_qualifier_code"] <- ""
htc %>%
filter(dmr_value_qualifier_code == "<") %>%
select(parameter_desc, dmr_value_qualifier_code, dmr_value_nmbr, nodi_desc)Flipping through the above table, it appears we have some kind of number listed in the ‘dmr_value_nmbr’ field for all the non-detect toxics which is what we want.
Aggregate Composite Parameters
In some cases, water quality objectives are expressed in terms of a composite parameter, like total PCBs, dioxins, or polyaromatic hydrocarbons. Sometimes these parameters are reported to the regulator in terms of their subconstituents or congeners instead of in terms of their total composite parameter. For example, let’s examine Hangtown Creek’s PCB data:
htc %>%
filter(stringr::str_detect(string = parameter_desc, pattern = "PCB"))In this case, PCBs were monitored once during the period of interest using seven PCB congeners. Under California’s implementing provisions, one would composite this data by adding together all the detected congener values on a given date. If none of the congeners were detected, then total PCB value would be reported as non-detect with a minimum detection limit equal to that of the lowest of minimum detection limits for the congeners. Let’s composite the total PCB results.
htc %>%
filter(stringr::str_detect(string = parameter_desc, pattern = "PCB")) %>%
group_by(monitoring_period_end_date) %>%
summarise(parameter_desc = "PCBs",
dmr_value_qualifier_code = if_else(n() == sum(dmr_value_qualifier_code == "<"), "<", ""),
dmr_value_nmbr = if_else(dmr_value_qualifier_code == "<",
min(dmr_value_nmbr, na.rm = TRUE),
max(dmr_value_nmbr[dmr_value_qualifier_code == ""],
na.rm = TRUE))) %>%
select(parameter_desc, monitoring_period_end_date, dmr_value_qualifier_code,
dmr_value_nmbr)## # A tibble: 1 x 4
## parameter_desc monitoring_period_end_da~ dmr_value_qualifier_c~ dmr_value_nmbr
## <chr> <date> <chr> <dbl>
## 1 PCBs 2017-04-30 < 0.001Using Data Exploration to Check for Suspicous Data
Let’s look at time-series scatter plots for hardness, lead, mercury, and zinc and see if anything pops out. Since we have a fair amount of data for lead, mercury, and zinc, chances are they have had reasonable potential for these parameters in the past or TMDLs are applicable. We will include hardness since it is used for assessing hardness-based metals toxicity.
htc %>%
filter(parameter_desc %in% c("Hardness, total [as CaCO3]",
"Zinc, total recoverable",
"Lead, total recoverable",
"Mercury, total recoverable")) %>%
ggplot(.) + geom_point(aes(x = monitoring_period_end_date,
y = dmr_value_nmbr,
col = dmr_value_qualifier_code)) +
facet_wrap(vars(parameter_desc), nrow = 4, scales = "free_y") +
labs(x = "", y = "Concentration")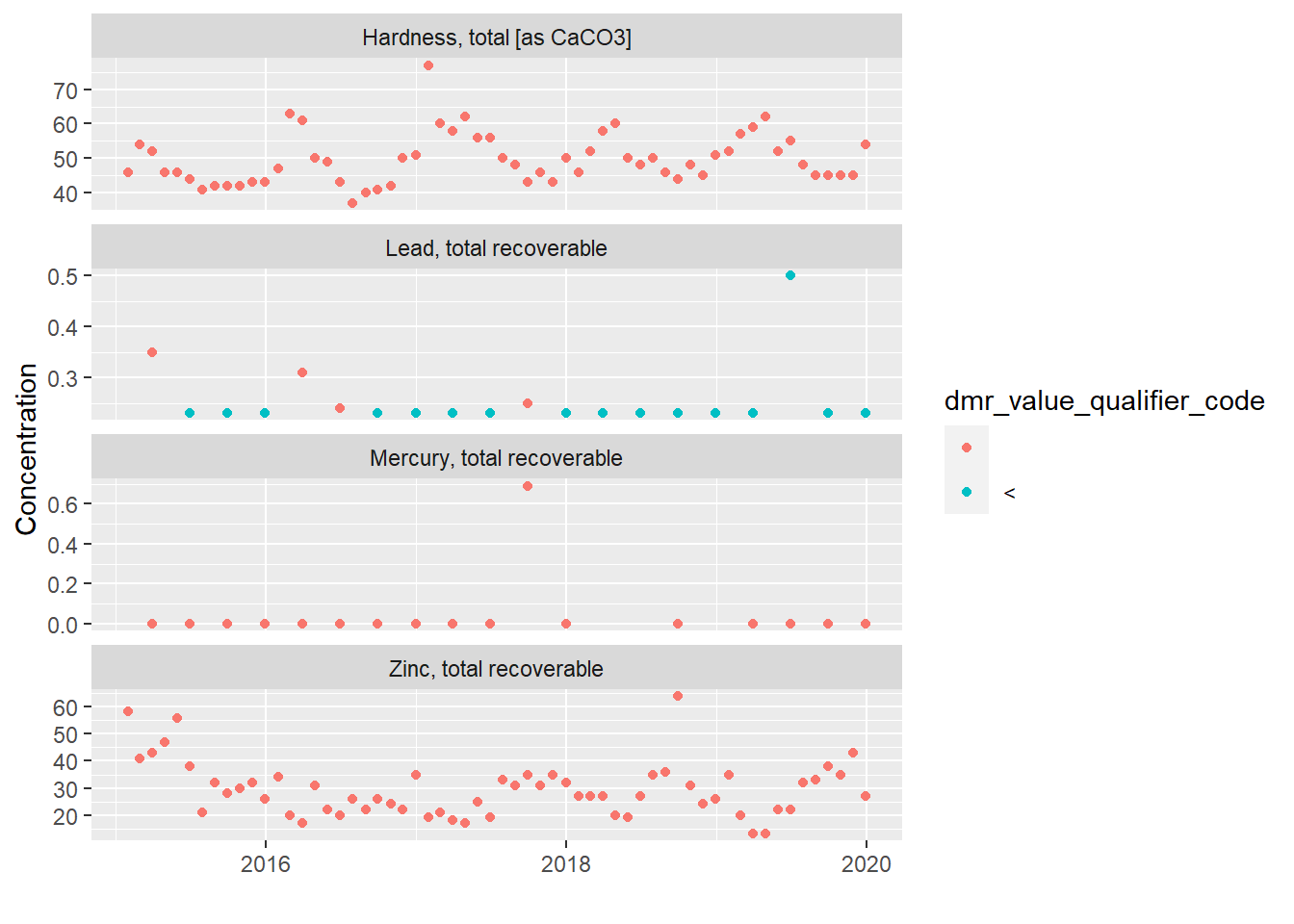
The 3rd quarter 2017 mercury result definitely stands out and merits further investigation. It is three orders of magnitude greater than all the other mercury results which immediately makes me wonder if its a decimal place typo. While the error appears obvious, I would need to confirm this suspicion before discounting the reported value. I would do this by contacting the permittee and either asking them to check their original lab sheet or to email a copy of the lab sheet so I can check it myself. “Outlier” datapoints should never be discarded unless it can be confirmed they are the result of measurement error or data entry error.
The lead measurement in 2nd quarter of 2019 has a different MDL than was used in other quarters which is a little odd but not obviously wrong, depending on the method sensitivity standards applicable to the plant.
Summarize the Data
Finally, we will summarize the data. Before doing this, we will prune our data down to the pollutants of concern. The pollutants of concern will depend on the effective water quality standards and associated implementing provisions in effect in the permittee’s jurisdiction. However, for this exercise we will assume the pollutants of concern are copper, heptachlor, lead, mercury, and zinc.
To perform and RPA in California, one would take the maximum observed concentration as the maximum effluent concentration (MEC). Under the TSD approach used in other jurisdictions, one would take the number of samples for a given constituent and the maximum observed concentration to project a MEC at a specified confidence interval. For this exercise, we will summarize the information needed for the TSD (though we won’t actually follow-through on projecting an MEC).
cv <- function(qual, result) {
new_result <- c(result[which(qual == "")], result[which(qual == "<")] * 0.5)
sd(new_result, na.rm = TRUE) / mean(new_result, na.rm = TRUE)
}
results <- htc %>%
filter(parameter_desc %in% c("Copper, total recoverable",
"Lead, total recoverable", "Mercury, total recoverable",
"Zinc, total recoverable", "Heptachlor")) %>%
group_by(parameter_desc) %>%
summarise(n = n(),
qual = if_else(n() == sum(dmr_value_qualifier_code == "<"), "<", ""),
result = if_else(qual == "<",
min(dmr_value_nmbr, na.rm = TRUE),
max(dmr_value_nmbr[which(dmr_value_qualifier_code != "<")], na.rm = TRUE)))
knitr::kable(results)| parameter_desc | n | qual | result |
|---|---|---|---|
| Copper, total recoverable | 1 | 3.1000 | |
| Heptachlor | 1 | < | 0.0018 |
| Lead, total recoverable | 20 | 0.3500 | |
| Mercury, total recoverable | 20 | 0.6900 | |
| Zinc, total recoverable | 60 | 64.0000 |
Putting it All Together
In summary, these were the data cleaning steps we followed:
We identified the pollutants of concern in the dataset and where they were measured (e.g., the effluent, the upstream and downstream receiving water).
We cleaned up the structure of the data (e.g., converting dates from a “character” class to a “date” class, converting concentration values to a “numeric” class).
Incomplete data was identified and retained for further investigation & follow-up. Examples of incomplete data include data missing units, or missing measurement values/MDLs.
We identified the data which represented single-sample measurements typically used in RPAs. The rest of the data–statistical aggregate values (average monthly, average weekly, etc.) which are computed based on the real measurements, and mass-flux (lb/day) values–were filtered out and discarded.
We processed and aggregated composite parameters based on their congeners/sub-constituents using PCBs as an example.
Compare the data present with what’s required to be collected in the permittee’s monitoring and reporting program. Is everything present and accounted for?
Suspicious data, like the high mercury measurement, was identified and flagged for further investigation.
We summarized the data.
Looking back on it, one thing a new permit writer should observe and take away is data cleaning involves lots of filtering, looking around, and thinking critically about what one sees within the data. While the steps above are a not-bad framework, data cleaning is more art than science and each project will be different. For example, one doesn’t typically summarise the MECs for Pacific Ocean discharge RPAs in California–instead one would need to combine the detected observations and the MDLs of non-detects for entry into California’s RPCalc software.
Regardless of the specifics, the key to success is to keep your eyes peeled, apply critical thinking skills, know the data format needed for the RPA, and to be thorough.
Tips
Always retain a copy of the raw data without any edits.
Compare the data collected with requirements listed in the permittee’s monitoring and reporting program–do you have all the data you are supposed to have for the RPA?
Look for missing information and, if it has the potential to change RPA results or limits, make certain to obtain it from the permittee or other sources.
Don’t discard “outlier” data unless there is a well-articulated and persuasive reason to do so. For RPA’s, one should investigate the authenticity of unusually large values but not discard them without proof that they are the result of some lab or data entry error.